Writing a Narrative¶
This documentation will walk us through constructing a narrative from scratch, based on this example (nextstrain.org). If you run into any bugs, please get in contact with us (email).
Step 1: Get the Underlying Datasets¶
You can skip this step if you have generated your own dataset, but for the purposes of this tutorial we will need the mumps dataset.
mkdir datasets
curl http://data.nextstrain.org/mumps_na.json --compressed -o datasets/mumps_na.json
You should now be able to visualise the dataset (without any narrative functionality) via auspice view --datasetDir datasets
Step 2: Start a Simple Narrative¶
We’re going to start by creating a narrative with only one page (the title page).
mkdir narratives
touch narratives/example.md
Open up narratives/example.md and start by pasting in the following YAML frontmatter, which is used to define some basic set-up for the narrative and create the first narrative page.
---
title: Example narrative tutorial
authors: "your name"
authorLinks: "url, twitter link, mailto etc"
affiliations: "your affiliation"
date: "August 2018"
dataset: "http://localhost:4000/mumps/na?d=tree"
abstract: "This narrative is going to test the potential of the Auspice narrative functionality using the publicly available North American mumps dataset."
---
The really important bit here is the dataset line – this references the dataset we downloaded above.
(A current limitation of narratives is that they cannot change the dataset.)
Step 3: View the Narrative so Far¶
We don’t really have much, but we can still load it up in Auspice and check it’s working as expected. If it is, we will see a narrative with only one page.
auspice view --datasetDir datasets/ --narrativeDir narratives/
And you should see something like this at localhost:4000/narratives/example
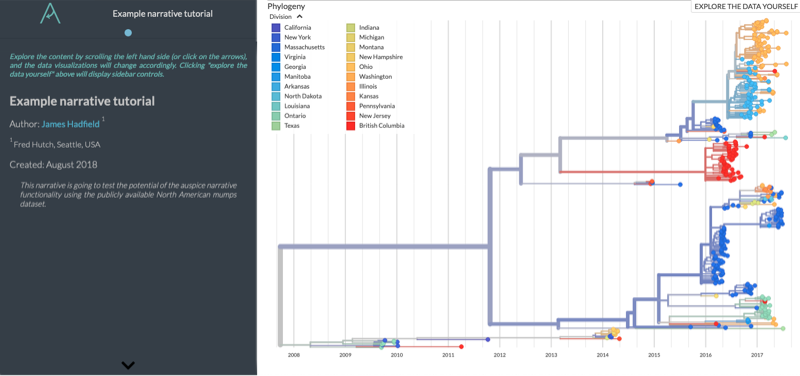
Step 4: Adding a Paragraph¶
Each “paragraph” or page of the narrative is made up of a section of Markdown starting with a h1 heading, which is a link with text beneath. It is these h1 headings that define when we have a new narrative pararaphs.
If you’re new to Markdown, take a look at this page to get started.
The heading is itself a link that defines the view of Auspice at that time. You may have noticed that as you interact with Auspice – for this example at localhost:4000/mumps/na – then the URL changes. For instance, if we use the sidebar to toggle “off” the tree and entropy, just keeping the map then you’ll see that the URL has changed to localhost:4000/mumps/na?d=map. We’re going to use this functionality to “save” the view of Auspice into the Markdown file, so that the narrative knows what view to show.
Let’s use this knowledge to make a page in the narrative which switches to just show the map. Add the following text to the narrative Markdown file:
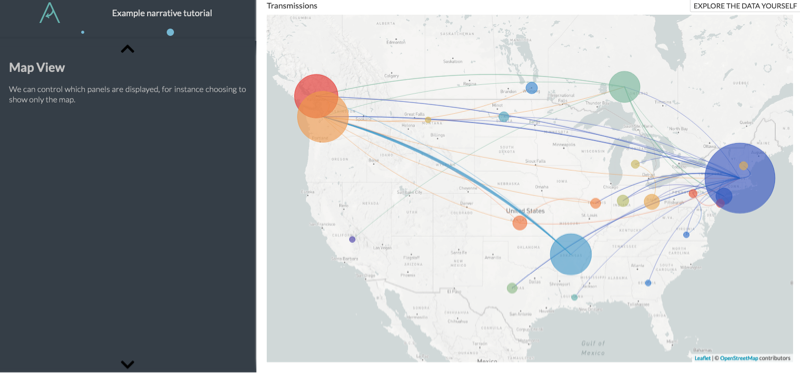
# [Map View](http://localhost:4000/mumps/na?d=map)
We can control which panels are displayed, for instance choosing to show only the map.
Now, we should have a second page available in the narrative:
Step 5: Adding More Paragraphs¶
We’re going to use the same iterative technique to add more pagragraphs:
Use Auspice (localhost:4000/mumps/na) to change the view as you desire
Copy this URL and create a h1 header in the markdown file
Write some text in the Markdown to form the narrative paragraph
Repeat until happy 😁
You can see the contents of the Markdown file behind the example (nextstrain.org) that we’re basing this tutorial on here. You can use this markdown file as inspiration for creating your own paragraphs, or just copy and paste the content! For reference, here are three paragraphs from that file:
# [Phylogenies](http://localhost:4000/mumps/na?d=tree&dmax=2014-07-14&dmin=2012-03-30&p=full)
Here we've returned to the tree and begun slicing time.
# [Explore clock signal](http://localhost:4000/mumps/na?d=tree&l=clock&p=full)
Different tree layouts are possible, this one shows the temporal
divergence vs. inferred substitutions to see the presence of a
constant clock signal.
# [Mutations I](http://localhost:4000/mumps/na?c=gt-SH_22&d=tree,entropy&p=full)
Here we have coloured the tree according to a single mutation (residue 22 in
the SH gene) where there are two variants present in this dataset -- yellow tips
have a Methionine (`M`) at this position, while aqua nodes indicate Isoleucine (`I`).